【math】Hiden Markov Model 隐马尔可夫模型了解
文章目录
- Introduction to Hidden Markov Model
- Introduction
- Markov chain
- Hidden Markov Model(HMM)
- Three Questions
- Q1: evaluate problem -- Forward algorithm
- Q2: decode problem -- Viterbi algorithm
- Q3: learn problem -- Baum-Welch algorithm
- Application
Introduction to Hidden Markov Model
Introduction
- Markov chains were first introduced in 1906 by Andrey Markov
- HMM was developed by L. E. Baum and coworkers in the 1960s
- HMM is simplest dynamic Bayesian network and a directed graphic model
- Application: speech recognition, PageRank(Google), DNA analysis, …
Markov chain
A Markov chain is “a stochastic model describing a sequence of possible events in which the probability of each event depends only on the state attained in the previous event”.
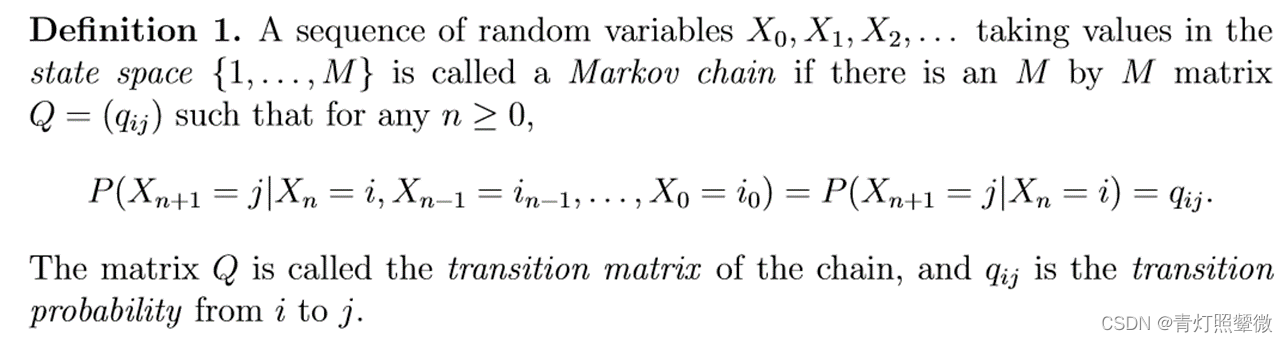
Space or time can be either discrete(𝑋_𝑡:t=0, 1, 2,…) or continuous(𝑋_𝑡:t≥0). (we will focus on Markov chains in discrete space an time)
Example for Markov chain:
- transition matrix 𝑄 :
- 5-step transition matrix is 𝑄^5 :

Hidden Markov Model(HMM)
HMM is a statistical Markov model in which the system being modeled is assumed to be a Markov process with unobserved (i.e. hidden) states.
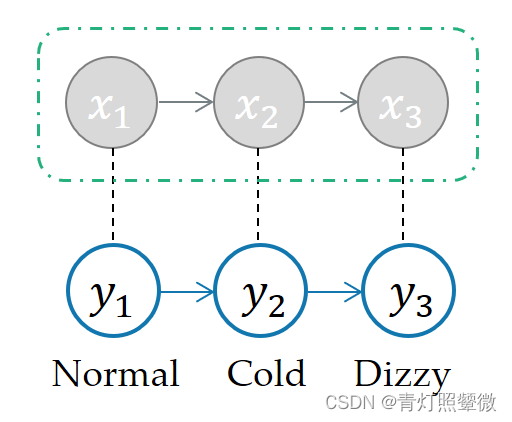
State: x = (x1, x2, x3) ;
Observation: y = (y1, y2, y3);
Transition matrix: A = (aij);
Emission matrix: B = (bij)
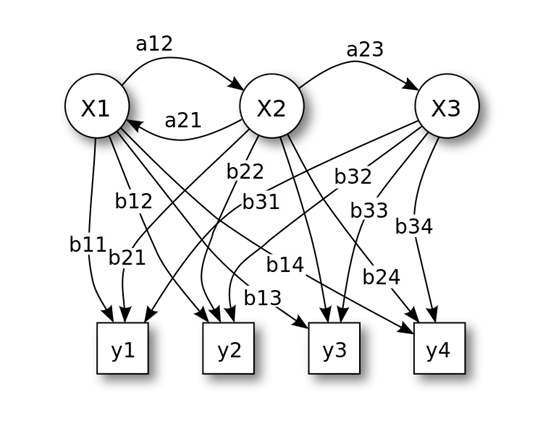
Example:

Three Questions
- Given the model 𝜆=[𝐴, 𝐵,𝜋], how to calculate the probability of producing the observation 𝒚={𝑦1_11,𝑦2_22,…,𝑦𝑛_𝑛n | 𝑦𝑖_𝑖i∈𝑂}? In other words, how to evaluate the matching degree between the model and the observation ?
- Given the model 𝜆=[𝐴, 𝐵,𝜋] and the observation 𝒚={𝑦1_11,𝑦2_22,…,𝑦𝑛_𝑛n| 𝑦𝑖_𝑖i∈𝑂}, how to find most probable state 𝒙={𝑥1_11,𝑥2_22,…,𝑥𝑛_𝑛n |𝑥𝑖_𝑖i∈𝑆} ? In other words, how to infer the hidden state from the observation ?
- Given the observation 𝒚={𝑦1_11,𝑦2_22,…,𝑦𝑛_𝑛n | 𝑦𝑖_𝑖i∈𝑂}, how to adjust model parameters 𝜆=[𝐴, 𝐵,𝜋] to maximize to the probability 𝑃(𝒚│𝜆) ? In other words, how to train the model to describe the observation more accurately ?
Q1: evaluate problem – Forward algorithm
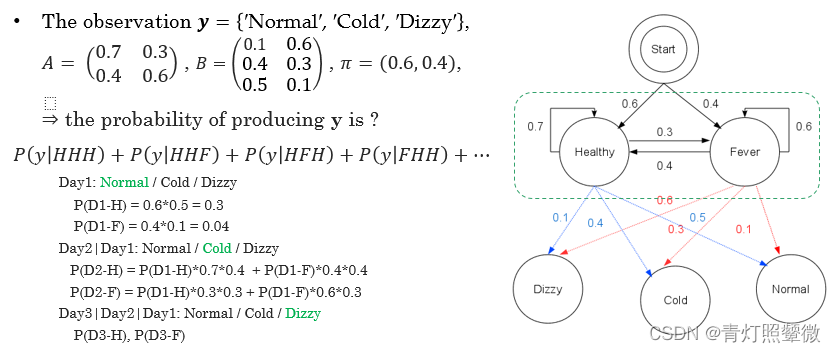
Q1: how to evaluate the matching degree between the model and the observation ? ( forward algorithm)
- the probability of observing event 𝑦1_11: 𝑃𝑖0_{𝑖0}i0 = 𝑃𝑖_𝑖i (𝑂=𝑦1{_1}1) = 𝜋𝑖𝑏1𝑖_{𝑖}𝑏_{1𝑖}ib1i
- the probability of observing event 𝑦𝑗+1_{𝑗+1}j+1 (𝑗≥1): 𝑃𝑖𝑗_𝑖𝑗ij=𝑏𝑗,𝑖+1_{𝑗,𝑖+1}j,i+1 ∑𝑘_𝑘k𝑃𝑖,𝑗−1_{𝑖,𝑗−1}i,j−1 𝑎𝑖𝑗_{𝑖𝑗}ij
- 𝑃(𝒚)=∑𝑘_{𝑘}k 𝑃𝑖𝑗_{𝑖𝑗}ij∗𝑎𝑗0_{𝑗0}j0
Q2: decode problem – Viterbi algorithm
Q2: how to infer the hidden state from the observation ? (Viterbi algorithm)
Observation 𝒚=(𝑦1_11, 𝑦2_22,…, 𝑦𝑇_𝑇T), initial prob. 𝝅=(𝜋1_11,𝜋2_22,…, 𝜋𝐾_𝐾K), transition matrix 𝐴, emission matrix 𝐵.
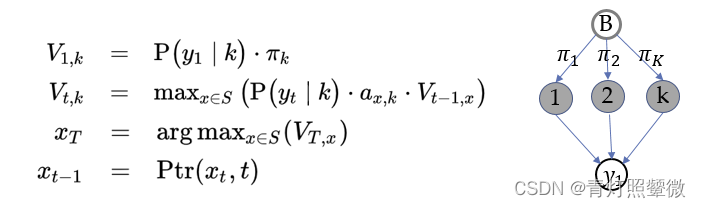
Viterbi algorithm(optimal solution) backtracking method; It retains the optimal solution of each choice in the previous step and finds the optimal selection path through the backtracking method.
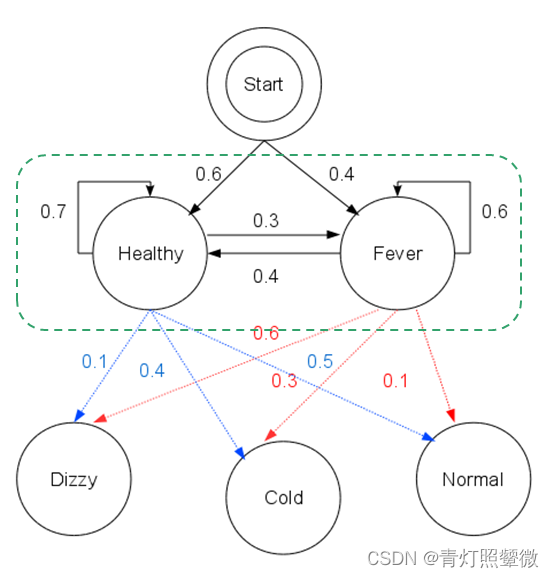
Example:
- The observation 𝒚={“′Normal′, ′Cold′, ′Dizzy′” } , 𝜆=[𝐴, 𝐵,𝜋], the hidden state 𝒙={𝑥1_11,𝑥2_22,𝑥3_33}= ?
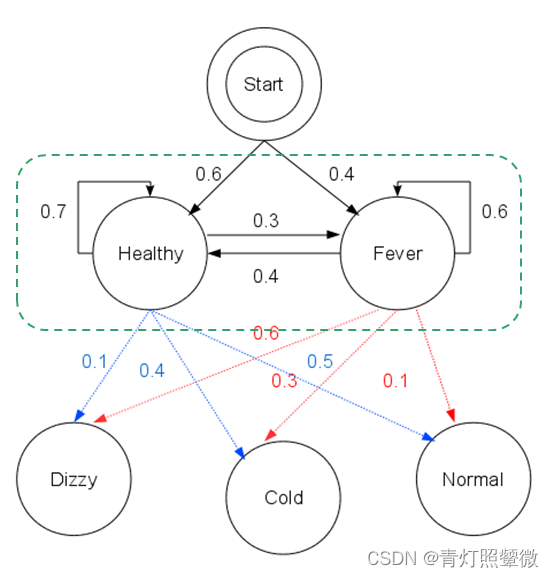
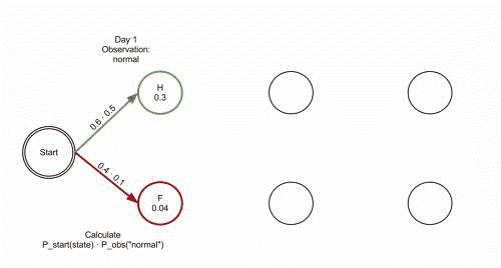
Q3: learn problem – Baum-Welch algorithm
Q3: how to train the model to describe the observation more accurately ? (Baum-Welch algorithm)
The Baum–Welch algorithm uses the well known EM algorithm to find the maximum likelihood estimate of the parameters of a hidden Markov model given a set of observed feature vectors.
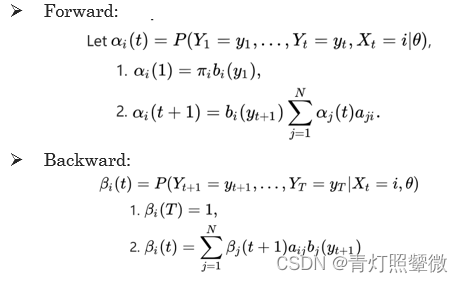
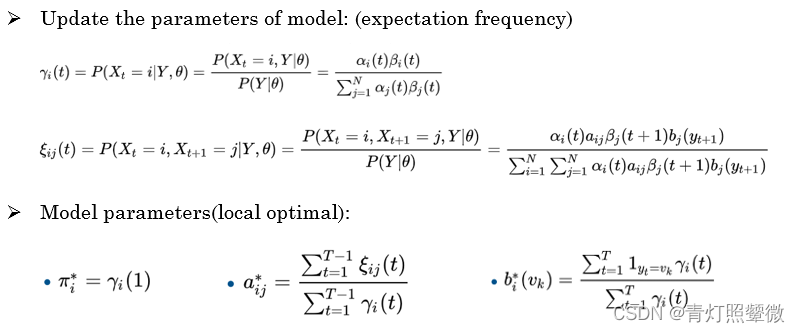
Baum–Welch algorithm(forward-backward alg.) It approximates the optimal parameters through iteration.

- (1) Likelihood function :
𝑙𝑜𝑔𝑃(Y,𝐼|𝜆) - (2) Expectation of EM algorithm:
𝑄(𝜆,𝜆 ̂ )= ∑𝐼_𝐼I 𝑙𝑜𝑔𝑃(𝑌,𝐼|𝜆)𝑃(𝑌,𝐼|𝜆 ̂ ) - (3) *Maximization of EM algorithm:
max 𝑄(𝜆,𝜆 ̂ )
use Lagrangian multiplier method and take the partial derivative of Lagrangian funcition。
Application
CpG island:
- In the human genome wherever the dinucleotide CG occurs, the C nucleotide is typically chemically modified by methylation.
- around the promoters or ‘start’ regions
- CpG is typically a few hundred to a few thousand bases long.


three questions: of CpG island: - Given the model of distinguished the CpG island, how to calculate the probability of the observation sequence ?
- Given a short stretch of genomic sequence, how would we decide if it comes from a CpG island or not ?
- Given a long piece of sequence, how would we find the CpG islands in it?
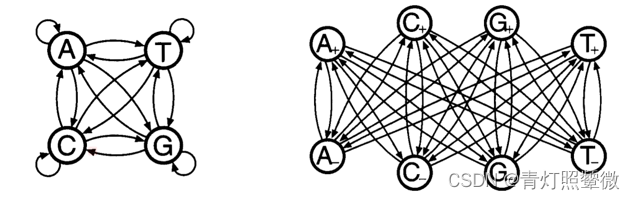
reference:
- Markov chain Defination
- A Revealing Introduction to Hidden Markov Models
- Top 10 Algorithms in Data Mining
- An Introduction to Hidden Markov Models for Biological Sequences
- python-hmmlearn-example
- Viterbi algorithm
- Baum-Welch blog
- Biological Sequence Analysis. Probabilistic Models of Proteins and Nucleic Acids. R. Durbin, S. Eddy, A. Krogh and G. Mitchison
未完待续…